WaveThresh Help
wave.band
Posterior credible intervals for wavelet regression
DESCRIPTION
Computes posterior credible intervals for an unknown regression curve.
USAGE
function(data = 0, alpha = 0.5, beta = 1., filter.number = 8, family =
"DaubLeAsymm", bc = "periodic", dev = var, j0 = 3., plotfn = T,
retvalue = T, n = 128, type = "data", rsnr = 3)
REQUIRED ARGUMENTS
Either data or a value of type other than
"data" must be supplied.
-
data
- If
type="data", then data should be a
vector of data. The length of the vector should be a power of two not
greater than 1024.
-
type
- Either
type="data", in which case a vector of data
should be supplied, or type should specify a standard
test function and wave.band will generate a test data set
via a call to test.data.
Permissible values for type are "blocks",
"bumps", "doppler", "heavi", or "ppoly"; see the documentation for test.data for more details.
OPTIONAL ARGUMENTS
-
alpha, beta
- Hyperparameters which determine the priors placed on the wavelet
coefficients. Both
alpha and beta take
positive values; see Abramovich,
Sapatinas, & Silverman (1998) or Chipman & Wolfson (1999) for more
details on selecting alpha and beta.
-
filter.number
- A parameter relating to the smoothness of wavelet that you
want to use in the decomposition.
-
family
- Specifies the family of wavelets to be used. Two popular options
are "DaubExPhase" and "DaubLeAsymm" but see the help for filter.select
for more possibilities.
-
bc
- Specifies the boundary handling. If
bc="periodic" the
default, then the function you decompose is assumed to be
periodic on it's interval of definition. Other boundary
options exist, but are currently unsupported for this function.
-
dev
- This argument supplies the function to be used to compute the
spread of the absolute values coefficients. The function supplied must
return a value of spread on the variance scale (i.e. not standard
deviation) such as the
var() function. A popular, useful
and robust alternative is the madmad
function.
-
j0
- The primary resolution level; used in assessing the universal threshold
which is used in the empirical Bayes estimation of hyperparameters.
-
plotfn
- If
plotfn=T, wave.band draws the noisy
data, the BayesThresh function estimate,
and pointwise 99 percent credible intervals for the regression
function. If the value of type is not
"data", then the true function will also be plotted.
-
retvalue
- If
retvalue=T, then a lengthy list of results will
be returned. Note that if both plotfn and
retvalue are set to F, then
wave.band will return no results whatsoever.
-
n
- If
type is not "data", then a data vector
of length n will be generated; note that n
should be a power of two not greater than 1024.
-
rsnr
- If
type is not "data", then the data
vector generated will have root signal-to-noise ratio as specified by
rsnr.
VALUE
If retvalue=F, the value returned by
wave.band is NULL. Otherwise,
wave.band returns a list with the following components:
-
data
- The data vector which has been analysed.
-
cumulants
- A list containing four vectors named one, two, three, and four.
Vector one contains the first cumulants of the regression function
estimate, vector to the second cumulants and so on.
-
Kr.wd
- A list of four wd
objects. These contain the first to fourth cumulants of the
wavelet coefficients, as well as recording the wavelet used in the
decomposition.
-
bands
- A list containing pointwise upper and lower credible limits for the
regression function estimate for nominal coverage rates 80, 90, 95 and
99 percent. The widths of the credible intervals is also included.
The vectors are named with "l", "u", and "w" indicating lower limits,
upper limits, and intervals widths, while "80", "90", "95", and "99"
refer to the nominal coverage rate.
-
param
- A record of parameters in the call to
wave.band.
SIDE EFFECTS
If plotfn=T, results are plotted on the current graphics
device.
DETAILS
This function implements the WaveBand method of Barber, Nason, & Silverman (2001) to
compute posterior credible intervals for a regression function. The
credible intervals are found by approximating the posterior
distribution of the estimated regression curve at each design point.
A mixture prior with two components (a zero-mean normal and a point
mass at zero) is placed on each wavelet coefficient and updated by the
data to give the posteriors for the wavelet coefficients. This is the
same prior used by Abramovich,
Sapatinas, & Silverman (1998) in their BayesThresh method,
implemented in the function BAYES.THR.
The cumulants of these posteriors are computed and stored in the wd
objects returned by wave.band as Kr.wd.
These are summed to give the posterior cumulants of the regression
curve, which are used to fit a Johnson distribution (Johnson, 1949), using the algorithm
of Hill, Hill, & Holder (1976).
Percentage points of these distributions are computed by the algorithm
of Hill (1976) and give the credible
intervals themselves.
Code to implement the algorithms by Hill
(1976) and Hill, Hill, & Holder
(1976) were obtained from the StatLib archive.
RELEASE
3.9.7
SEE ALSO
BAYES.THR
plot.wb
power.sum
test.data
EXAMPLES
#
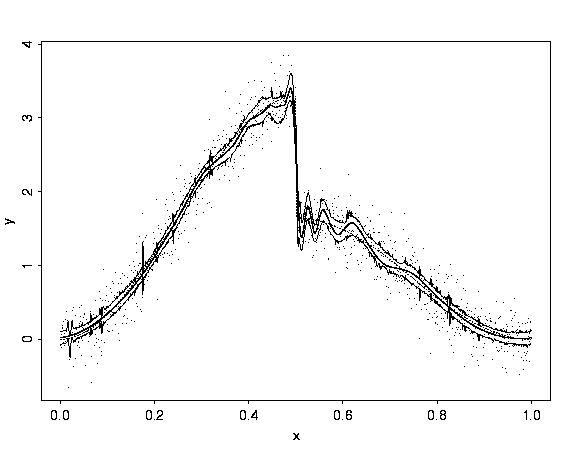
# First, look at the piecewise polynomial example.
#
# This plot and the plots for the smooth example below show
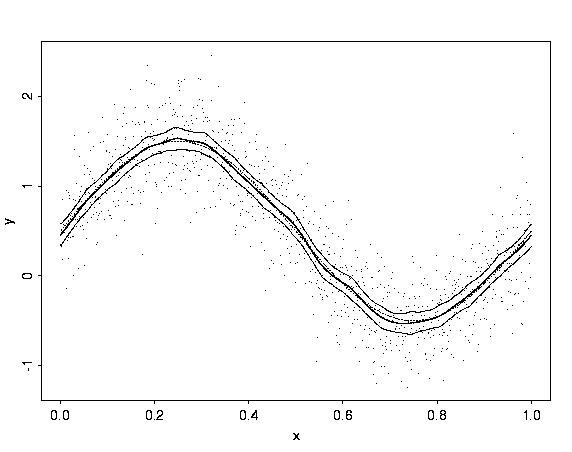
# the data as points, the BayesThresh estimate (thick line),
# pointwise 99 percent credible intervals (thin lines), and
# the true function (dotted thin line).
#
ppoly.wb <- wave.band(type = "ppoly", n = 1024, rsnr=4)
#
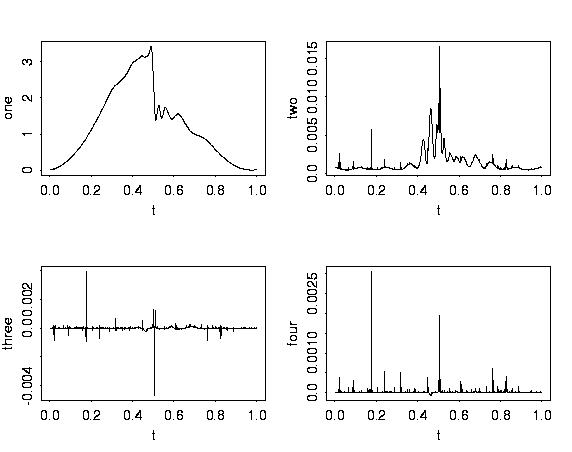
# Plotting the cumulants shows that there are significant
# third and fourth cumulants in some places.
#
t <- (1:1024)/1024
plot(t, ppoly.wb$cumulants$one, type="l", xlab="t", ylab = "one")
plot(t, ppoly.wb$cumulants$two, type="l", xlab="t", ylab = "two")
plot(t, ppoly.wb$cumulants$three, type="l", xlab="t", ylab = "three")
plot(t, ppoly.wb$cumulants$four, type="l", xlab="t", ylab = "four")
#
# Now consider how much difference the prior can make.
# Consider a smooth example, first using the default prior,
# and then using a smoother prior.
#
gs <- sin(2*pi*t) + 2*(t - 0.5)^2
gs.noisy <- gs + rnorm(n=1024, sd=sqrt(var(gs))/2)
gs.wb1 <- wave.band(data=gs.noisy)
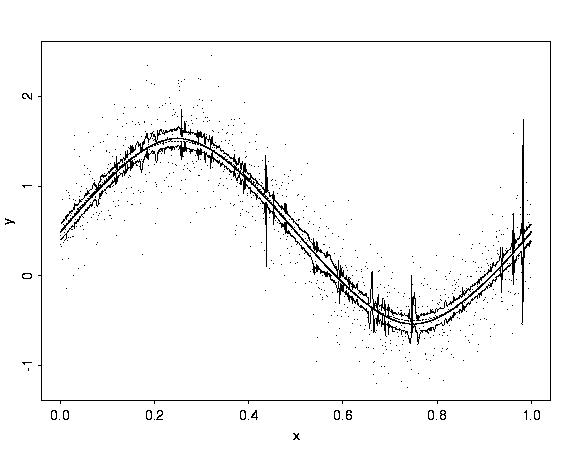
gs.wb2 <- wave.band(data=gs.noisy, alpha=4, beta=1)