About fineSTRUCTURE
fineSTRUCTURE is a fast and powerful algorithm for identifying population structure using dense sequencing data. By using the output of ChromoPainter as a (nearly) sufficient summary statistic, it is able to perform model-based Bayesian clustering on large datasets, including full resequencing data, and can handle up to 1000s of individuals. Full assignment uncertainty is given.There is a Graphical User Interface (GUI) as well as a command line version, and the program runs on both Windows and Linux (Mac support may be possible).
A Stochastic optimization routine is available for performing faster EDA and dealing with larger datasets - see FAQ under "What if my dataset is too big for MCMC".
Download fineSTRUCTURE
Citation and further information
The correct citation is:- Lawson, Hellenthal, Myers, and Falush (2012), "Inference of population structure using dense haplotype data", PLoS Genetics, 8 (e1002453). (preprint and Supporting Information in a single PDF file rather than many individual files).
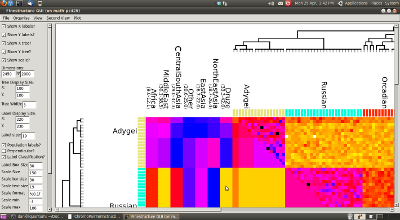
- Additional Supplementary material (the Whole World HGDP heatmap).
Full details of the process are given in that reference. You may also want to see an early overview poster which describes the broad approach.